Establishment and Maintenance of Subgenome Dominance in Allopolyploids
Many of the world's most important crops are polyploid or have recent polyploid origins, including canola, cotton, potato, maize, and wheat. In many cases, these polyploid species are formed through interspecific hybridization and whole genome duplication (WGD), which are termed allopolyploid. Because these species contain the full genomic compliment of two or more progenitor species, they have an expanded "tool-kit" of genes which can be used to enhance survival and in some cases expand into new niches.
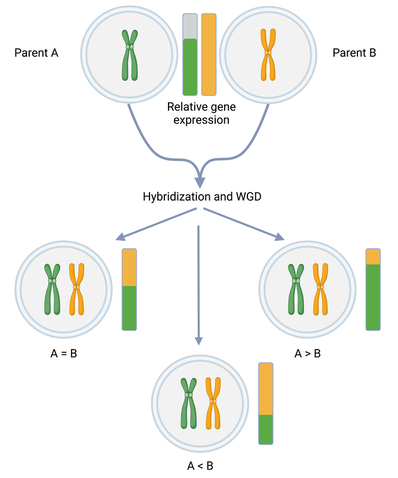
Hybridization and whole genome duplication are often observed to lead to imbalances in the resulting polyploid species. One such imbalance is expression-level-dominance, when one subgenome has heightened transcriptional activity (dominance) relative to the other(s). In fact, many polyploid crop species exhibit subgenome dominance at the expression-level. How this is established, and what role it might play in shaping traits of agronomic importance remains unclear. My research focuses on the potential factors affecting this dominance phenomenon and how they might be leveraged for crop improvement. Specifically, I am looking at the role of environmental effects, both before and after hybridization and WGD. I am approaching this question through a two-pronged approach of modifying the environment of diploids and conducting crosses and resynthesizing polyploids in various environments and also subjecting already established natural polyploids to different environments to determine whether environment can elicit shifts in dominance.
Development of Camelina as a Genomic Model System
The genus Camelina contains diploids, tetraploids, and hexaploids, and with relatively small genome sizes (~200-750Mb) these taxa represent promising tools for genomics and agricultural research. Due to the recent hybrid history of tetraploid and hexaploid species, and existence of their diploid progenitors in the wild, it is possible to resynthesize these hybrids as a route to expand genetic variation, specifically in the crop, C. sativa. The relatively short generation times and ease of propagation for many Camelina species sets them up well to be used as genetic model systems in a laboratory environment. However, several challenges and uncertainties are currently prohibiting this endeavor.
My works aims to introduce new high-quality and highly contiguous reference genomes for Camelina species. Without such genomic resources, much of the utility of the system is inaccessible. Further, I will generate efficient transformation protocols for wild Camelina species to accelerate their use in genetic research.
My works aims to introduce new high-quality and highly contiguous reference genomes for Camelina species. Without such genomic resources, much of the utility of the system is inaccessible. Further, I will generate efficient transformation protocols for wild Camelina species to accelerate their use in genetic research.
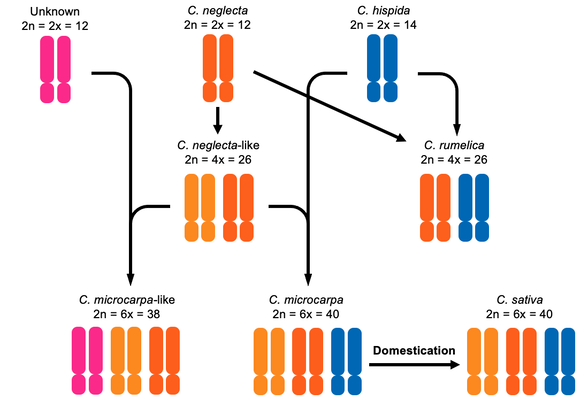
Evolutionary History of Camelina (Brassicaceae) and Origins of Gold-of-Pleasure
A crop for thousands of years in southwestern Asia and Europe, gold-of-pleasure (Camelina sativa) served many uses as a forage, oilseed crop for food and fuel, and for fiber. Approximately one-hundred years ago, cultivation of this ancient crop largely ceased throughout Europe and Asia, although the reasons for this are not well documented. Recently a resurgence of research has identified gold-of-pleasure as a suitable crop for the production of renewable biofuels, in particular for the aviation industries. However, little was known about the genus Camelina, and how the ancient oilseed gold-of-pleasure was domesticated. I study the evolutionary history of this group using phylogenetic, population genetic, and genomic techniques. In the process, I work through the long-standing taxonomic challenges in the genus and describe new variation that was previously unrecognized.